It’s been a long time since I wanted to test this Arch-based Linux distribution, XeroLinux. Finally, I found some time, and I first tested it in VirtualBox, and then I also installed that on one of my laptops.
What XeroLinux is about is summarized on its home page:
A Hobbyist Arch-Based Distro, built using ArcoLinux’s ALCI scripts that is meant to provide you with a stable base, making it simpler to install ArchLinux (Calamares) with a well optimized & Customized KDE Plasma, that you can shape YOUR way, the easy way… !
It’s an Arch-based distribution (not like Manjaro): it’s basically Arch with a few additional repositories that are mainly devoted to one of the main exciting features of XeroLinux, that is, a beautiful (good looking) KDE customized experience. And, although the “beauty is in the eye of the beholder”, XeroLinux KDE DE is indeed amazingly beautiful.
In this (long) post, I will summarize my overall experience of XeroLinux, starting from the installation to daily usage. Maybe it’s because I’m used to Arch, but I really like it!
Installation
The installation process starts, as usual, with a Live ISO. One of the fun things is that when you boot the Live image, the GRUB countdown emits a beep every second 🙂
When you get to the live environment, you can already understand the beauty of the KDE customizations (dock, global menus, blurring, etc.)
You’re then greeted by the XeroLinux Hello program:
If on a virtual machine, you can use one of the two buttons to fix the screen resolution. In general, you might want to “Update Arch Mirrorlist” right away. Then, just “Launch installer”.
The installer is based on the good old Calamares. If you’re already familiar with that, it will be easy! If not, it will not be that hard since the installer will ask you only the basic questions and will install all the default packages without further intervention.
Of course, the “Partitions” section is the critical one.
You probably wipe the whole (virtual) disk in a virtual machine. As usual, you can select a swap partition with hibernate. XeroLinux defaults to XFS, but I prefer BTRFS, which I’ve selected in my installations. As we’ll see later, the default layout of BTRFS subvolumes created by XeroLinux is also good!
When installed on my laptop, where I have several Linux installations, I chose the manual partitioning and manually created a new BTRFS partition while mounting another shared data partition. As usual, if you have an existing EFI partition, you must manually mount it in “/boot/efi”:
After reviewing the summary, it’s time to start the installation:
Do you see that green button on the bottom right? Click it, and you can see the log of the installer:
When the installation finishes, you get a message that the installation log can be found in the installed system:
The name of the LOG file is wrong. I opened a PR, https://github.com/xerolinux/calamares-config/pull/1. In any case, finding the LOG file in the /var/log directory in the installed system is easy.
Note: the first time I tried the installer on a virtual machine, Calamares crashed:
I had to start from scratch. The next time, I updated the mirrors before starting the installation, and the installation went fine. I can’t tell what went wrong the very first time.
The installed system
Let’s start with the impressions about the installed system.
The BTRFS subvolume layout looks good to me, or at least, that’s how I create the subvolumes myself in other distributions:
|
1 2 3 4 5 6 7 8 |
$ sudo btrfs su li / ID 256 gen 220 top level 5 path @ ID 257 gen 221 top level 5 path @home ID 258 gen 103 top level 5 path @root ID 259 gen 39 top level 5 path @srv ID 260 gen 106 top level 5 path @cache ID 261 gen 219 top level 5 path @tmp ID 262 gen 216 top level 5 path @log |
where, besides the standard @ and @home, we have @log, @tmp and @cache that correspond to the directories in /var, @root is for the /root folder, and @srv for /srv.
These are the mount options of the BTRFS subvolumes (in the /etc/fstab file):
|
1 |
defaults,noatime,noautodefrag,compress=zstd,discard=async,ssd |
I don’t like the “discard=async” in the file /etc/fstab. Since fstrim is enabled once a week, I prefer not to have “discard=async” so I edited /etc/fstab accordingly. From what I know, the “ssd” should not be necessary since BTRFS should recognize that it’s using an SSD. However, it shouldn’t harm.
Another thing I don’t like is that the journald log is not persisted on reboots. I reported that on the forum, https://forum.xerolinux.xyz/thread-184.html. Apparently, that’s not intended. I can fix this by editing /etc/systemd/journald.conf and changing the line
|
1 |
Storage=volatile |
to the default (you could simply remove the line, but I prefer to comment it to remember that the default is “auto”):
|
1 |
#Storage=auto |
Moreover, I had to delete the file /etc/systemd/journald.conf.d/volatile-storage.conf (which also contains the “Storage=volatile”). Reboot, and now the log stays there even after rebooting.
The home page mentions, “This distro is 90% ArchLinux & 10% custom.” Indeed, these are the additional packages https://github.com/xerolinux/xerolinux_repo/tree/main/x86_64. Besides a few pre-compiled packages (instead of the source versions of the same packages in the AUR repository), there are many packages about theming. I’d tend to say that XeroLinux is “very” Arch, and I like that.
The installed KDE
As I had already anticipated, the KDE desktop environment, customized by XeroLinux, looks really good. It uses many parts from the Lyan theme and Tela icons (I had already blogged about that), however, for the “Application Style”, it uses Lightly instead of Kvantum (as shown in the XeroLinux site and videos, you can easily switch to different themes, actually “rices”, provided by XeroLinux itself). I was already a fan of Lyan and Tela, so I liked the outcome.
You find a dock at the bottom, which is a Latte dock. (By the way, by default, the Latte dock is set to “Dodge Maximized”, which is not ideal for me: I prefer “Dodge Active”.) Also, the top panel is not a standard panel: it’s, again, a Latte panel. They are both transparent (blurred), and you have a global menu, a la macOS. I’m not a big fan of the global menu, but I’ll try to stick with that for the time being. There are a few useful widgets: in the bottom dock, you have the CPU usage, and in the top panel, you have the network usage.
For KDE applications, you don’t need to go to the menu if you use the shortcut “Ctrl + Alt + I,” which opens a global search on the menus of that application (the so-called HUD). For example, here’s the HUD for Dolphin:
Speaking of KDE Desktop effects, you have several effects enabled by default: “Slide back”, “Wobby Windows”, etc. Not the “Magic Lamp” effect, which is one of my favorites, but enabling that is easy.
Moreover, you get “blurred” in most places. It’s eye candy, and I like it 🙂 In that respect, as mentioned on the XeroLinux site, you need a powerful computer to deal with all these effects. On my Dell XPS 13, everything is smooth. Moreover, I don’t see such effects impacting power consumption when on battery.
Look at the blurred effect on the custom Konsole profile (which is set as the default one):
And what about the cool “ls” output? Well, that’s because “ls” is automatically aliased to exa, a modern replacement for ls.
Here are the blurred menus:
Lightly also can blur the Dolphin view, which is enabled by default. Unfortunately, since I’m using fractional scaling, that does not work.
By the way, Dolphin is already configured with the Dropbox plugin enabled. As soon as you install Dropbox, you have overlay decorations on the Dropbox folder.
Moreover, also the KDE drop-down terminal, Yakuake, is already configured and ready to use with F12.
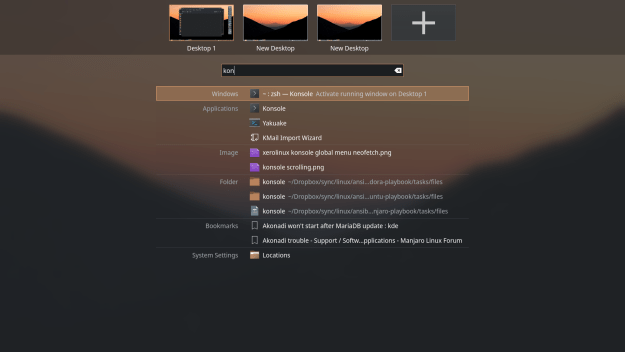
Speaking of searching, the custom “Application Menu” separates search results (files, i.e., “Desktop search”, “Settings”, “Applications”, etc.):
A thing that gave me a few headaches with Kalendar and Korganizer (e.g., for accessing Google Calendar accounts) is that the KWallet subsystem is DISABLED by default (that’s one intentionally). As a consequence, I wasn’t able to access any Google calendar without any feedback. However, it’s just a matter of enabling the KWallet subsystem. By the way, I have no problem with the KWallet subsystem enabled, despite what was described in the XeroLinux forum as the motivation for disabling the KWallet subsystem.
XeroLinux specific features
The Xero Hello program is also very useful! It provides several menus
The “Install Drivers”:
I particularly like the “Post-Install System Configuration”:
For example, it has a menu for switching the shell from BASH to ZSH with Oh My Zsh (including a few common plugins, e.g., completions, suggestions, and source highlight, not to mention the beautiful Powerlevel10k theme). It takes care of cloning Oh My Zsh, and its plugins and sets the shell for the current user:
Speaking of beauty, XeroLinux comes with a very good set of good-looking wallpapers! (see https://github.com/xerolinux/kde-wallpapers/tree/main/usr/share/wallpapers):
The “Update System Now” is also very powerful. It gives you a choice to update Everything (by relying on Topgrade): not only Pacman, AUR, and Flatpak packages but also fuwpd, Oh My Zsh and its plugins (if you installed that), and Docker images (if you have installed Docker).
Final thoughts
I like XeroLinux, and I like its BEAUTIFUL theming! It makes me want to use KDE again 🙂
Most of all, I think it is similar to EndeavourOS, which I also like: it’s basically Arch (not like Manjaro, which I evaluated in the past, but which somehow twisted Arch) with interesting customizations.